FMRI navigation
From ACL@NCU
Preprocessing
- These notes apply to 140.115.47.153
Creating alias
- Create alias of commands (or “shortcuts”), under the home directory, use the nano command to edit a configuration file:
nano .tschrc.aclexp
- Add to the bottom of the file:
alias <name_of_shortcut> <‘your command’>
- Replace contents within <> to suit your need. For example:
alias cdnav ‘cd /media/DATA1/navigation’
allows you to type just “cdnav” (without the quotes) to change directory from anywhere to /media/DATA1/navigation. You will have to reinitiate another instance of terminal for the new settings to be effective.
GUI interface
- You can initiate a graphic user’s interface (gui) of file manager by typing:
nautilus
- Or a gui editor by typing
gedit
- Note that when you initiate a gui in a terminal, you won’t be able to type new commands in that terminal. You can initiate another terminal from the SSH client gui.
Reconstructing rawdata into AFNI format
- Change working directory to the navigation folder
cdnav
- Change working directory to the script folder
cd scripts
- edit the participants list
nano subjlist.txt
- To 3D
python to3d_anat.py <numid>
-numid: numerical code of the participant.
python to3d_func.py <numid>
-numid: numerical code of the participant.
Creating stimulus time files
- Change working directory to the scripts folder
cd /media/DATA1/navigation/scripts
-Or just type “cd scripts”, if you have followed settings in previous slides to change directory to /media/DATA1/navigation
- Creat stimulus time files
python mkcond1D03.py <numid> <group id>
-numid: numerical code of the participant (001, 002, … etc)
-group id: group code of the participant (allo or ego )
Running Preprocessing & Single-subject GLM
- To carry out all preprocessing steps
-cutting out TR (pb00), outlier screening, normalization (pb01), realignment (pb02), spatial smoothing (pb03), and scaling (pb04)
./proc.prep <groupid> <initials+date>
-groupid: the participant’s group (allo/ego)
-initials+date: participant’s initials followed by date (e.g. LPS141109 ).
- To carry out single-subject (first level) GLM
./proc.glm <groupid> <numid> <initials+date>
-groupid: the participant’s group (allo/ego)
-numid: numerical code of the participant
-initials+date: participant’s initials followed by date (e.g. LPS141109 ).
Checking & saving results
- Change to group.allo folder
cdreva
-you need to create this alias
- Change to group.ego folder
cdreve
-you need to create this alias
- Change to folder of the participants you would like to see and save the results
./@ss_review_basic > review_<id> .txt
-id: name+date – LPS141109 -save results
./@ss_review_driver
-check brain images
Details of checking files
- Average outlier frac :
-notice if > 0.05
- Average motion :
-notice if > 1
- Fraction censored per run :
-notice if > 0.1
Details of checking images
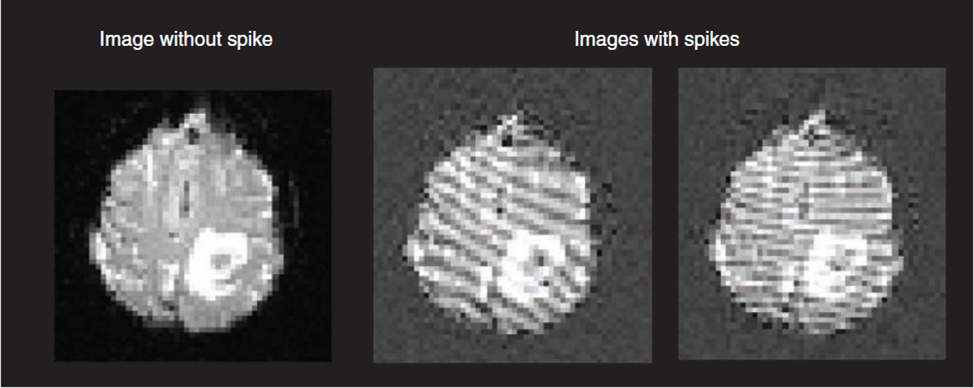
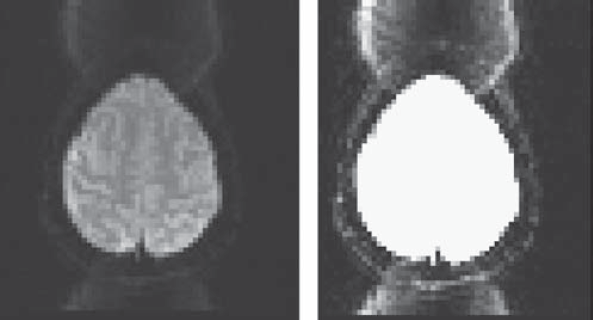
- Check whether there are spikes or ghosts from the brain image.
- Check ghosts with “pb00” files from afni
- Check spikes : change to folder of the participants
./@epi_review.<ID>
-ID: initials+date – LPS141109
-after checking a run, press enter on command window to go through the next run and so on.
Saving to Excel
- Save these three information to the excel worksheet ()
-Number TRs above not limit
-Number TRs above out limit
-Censor fraction